Filter Segments By Mask
SUMMARY
Filter Segments By Mask filters superpixels based on a mask.
Filter Segments By Mask keeps only superpixels that overlap with a specified mask region. This is useful for spatial filtering, such as keeping only superpixels within a region of interest.
Use this Skill when you want to filter superpixels based on spatial overlap with a mask.
The Skill
from telekinesis import cornea
result = cornea.filter_segments_by_mask(
image=image,
labels=labels,
mask=mask)Example
Input Image
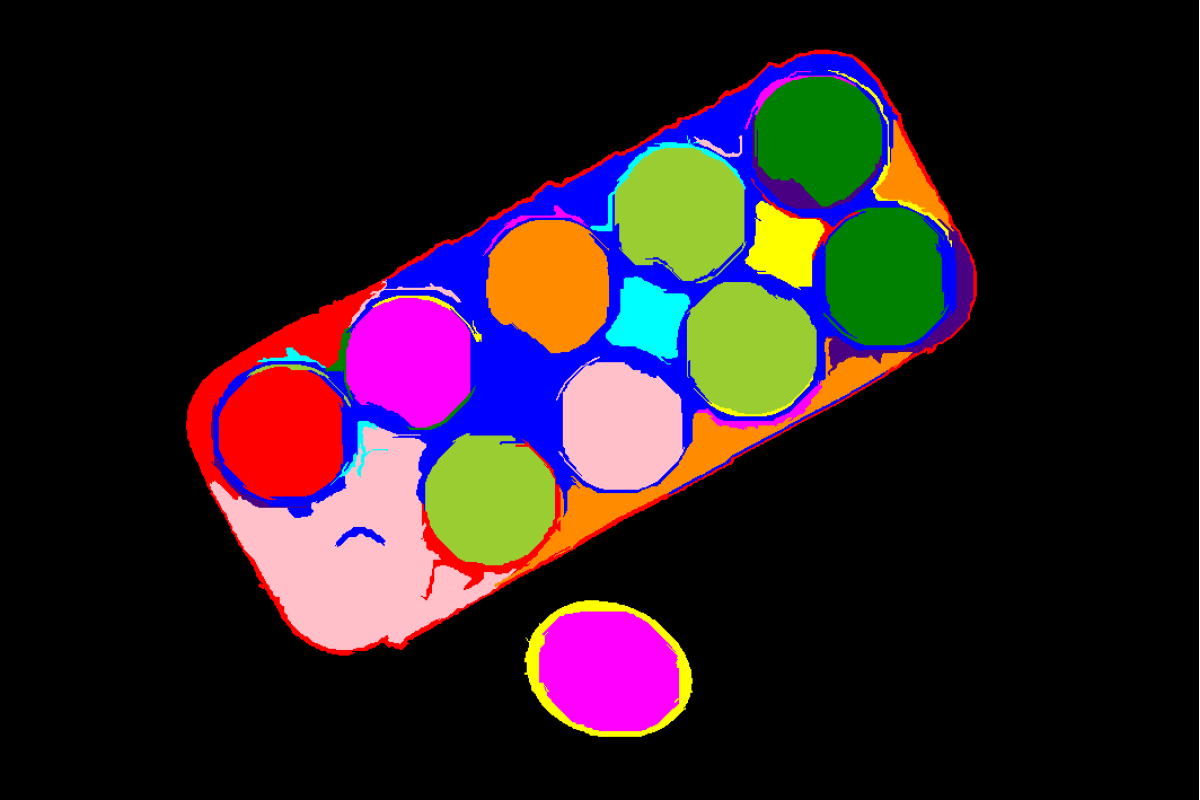
Original image with superpixels
Output Image
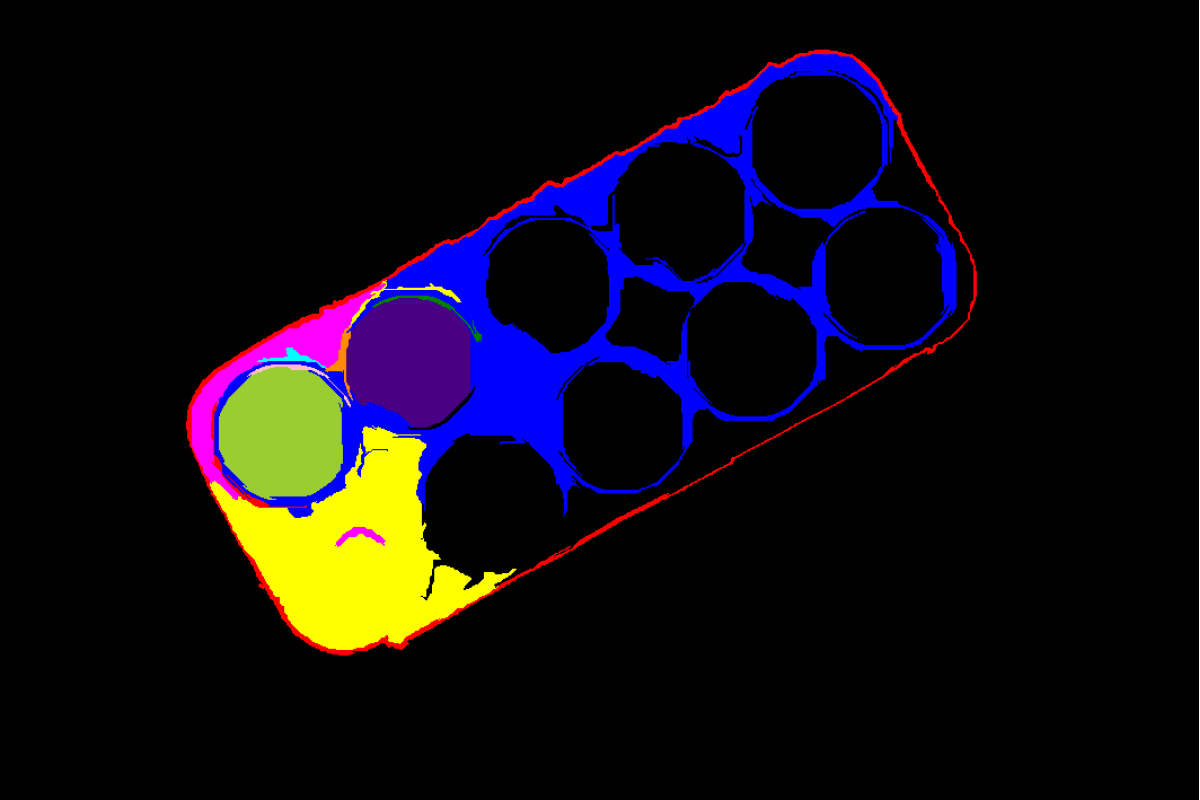
Filtered image - only superpixels within mask region shown
The Code
import pathlib
import numpy as np
from telekinesis import cornea
from datatypes import datatypes, io
DATA_DIR = pathlib.Path("path/to/telekinesis-data")
# Load image
filepath = str(DATA_DIR / "images" / "eggs_carton.jpg")
image = io.load_image(filepath=filepath)
# First, generate superpixels using Felzenszwalb
result_felzenszwalb = cornea.segment_image_using_felzenszwalb(
image=image,
scale=500,
sigma=1,
min_size=200,
)
# Extract superpixel labels
superpixel_labels = result_felzenszwalb["annotation"].to_dict()['labeled_mask']
superpixel_labels = datatypes.Image(superpixel_labels)
# Create a mask (left third of the image)
h, w, _ = image.to_numpy().shape
mask_np = np.zeros((h, w), dtype=np.uint8)
mask_np[:, :w//3] = 255
mask = datatypes.Image(image=mask_np, color_model='L')
# Filter superpixels based on mask
result = cornea.filter_segments_by_mask(
image=image,
labels=superpixel_labels,
mask=mask,
)
# Access results
annotation = result["annotation"].to_dict()
filtered_mask = annotation['labeled_mask']The Explanation of the Code
Filter Segments By Mask keeps only superpixels that have sufficient overlap with the specified mask region, effectively performing spatial filtering.
The code begins by importing the required modules, loading an image, and generating superpixels:
import pathlib
import numpy as np
from telekinesis import cornea
from datatypes import datatypes, io
DATA_DIR = pathlib.Path("path/to/telekinesis-data")
filepath = str(DATA_DIR / "images" / "eggs_carton.jpg")
image = io.load_image(filepath=filepath)
# Generate superpixels first
result_felzenszwalb = cornea.segment_image_using_felzenszwalb(
image=image,
scale=500,
sigma=1,
min_size=200,
)
superpixel_labels = result_felzenszwalb["annotation"].to_dict()['labeled_mask']
superpixel_labels = datatypes.Image(superpixel_labels)A mask is created to define the region of interest (left third of the image):
h, w, _ = image.to_numpy().shape
mask_np = np.zeros((h, w), dtype=np.uint8)
mask_np[:, :w//3] = 255
mask = datatypes.Image(image=mask_np, color_model='L')The filter parameters are configured:
result = cornea.filter_segments_by_mask(
image=image,
labels=superpixel_labels,
mask=mask,
)The function returns a dictionary containing an annotation object. Extract the mask as follows:
annotation = result["annotation"].to_dict()
filtered_mask = annotation['labeled_mask']Running the Example
Runnable examples are available in the Telekinesis examples repository. Follow the README in that repository to set up the environment. Once set up, you can run this specific example with:
cd telekinesis-examples
python examples/cornea_examples.py --example filter_segments_by_maskHow to Tune the Parameters
The filter_segments_by_mask Skill has 2 parameters:
labels (required):
- Label image from superpixel segmentation
- Units: Image object
- Each unique value represents a different superpixel
mask (required):
- Mask image defining the region of interest
- Units: Image object
- Non-zero pixels define the region to keep
- Should match the size of the image and labels
TIP
Best practice: Create the mask using other segmentation methods or user input. The mask should clearly define the spatial region of interest.
Where to Use the Skill in a Pipeline
Filter Segments By Mask is commonly used in the following pipelines:
- Region of interest filtering - Keep only superpixels in ROI
- Spatial filtering - Filter based on location
- Interactive segmentation - Filter based on user-selected region
- Post-processing - Refine superpixel segmentation spatially
A typical pipeline for spatial filtering looks as follows:
from telekinesis import cornea
from datatypes import datatypes, io
# 1. Load the image
image = io.load_image(filepath=...)
# 2. Generate superpixels
superpixel_result = cornea.segment_image_using_felzenszwalb(image=image, ...)
# 3. Extract labels
superpixel_labels = superpixel_result["annotation"].to_dict()['labeled_mask']
superpixel_labels = datatypes.Image(superpixel_labels)
# 4. Create or load mask (e.g., from user selection or detection)
mask = datatypes.Image(image=mask_np, color_model='L')
# 5. Filter by mask
result = cornea.filter_segments_by_mask(
image=image,
labels=superpixel_labels,
mask=mask,
)
# 6. Extract filtered mask
annotation = result["annotation"].to_dict()
filtered_mask = annotation['labeled_mask']Related skills to build such a pipeline:
segment_image_using_slic_superpixel: Generate superpixelsfilter_segments_by_area: Filter by areafilter_segments_by_color: Filter by color
Alternative Skills
| Skill | vs. Filter Segments By Mask |
|---|---|
| filter_segments_by_area | Filter by area instead of space. Use area for size-based filtering, mask for spatial filtering. |
| filter_segments_by_color | Filter by color instead of space. Use color for color-based filtering, mask for spatial filtering. |
When Not to Use the Skill
Do not use Filter Segments By Mask when:
- You need size-based filtering (Use filter_segments_by_area instead)
- You need color-based filtering (Use filter_segments_by_color instead)
- You don't have a mask (Create mask first or use other filtering methods)
TIP
Filter Segments By Mask is particularly useful for interactive applications where users can define regions of interest, or when you have prior knowledge about object locations.

